 Samples are stored in RNA later solution to preserve the nucleic acids.
Samples are stored in RNA later solution to preserve the nucleic acids. Samples are disintegrated in liquid nitrogen.
Samples are disintegrated in liquid nitrogen. A series of chemical procedures are used to extract and purify the nucleic acids.
A series of chemical procedures are used to extract and purify the nucleic acids. The genetic code of the strain in the sample is determined using the latest sequencing technology.
The genetic code of the strain in the sample is determined using the latest sequencing technology.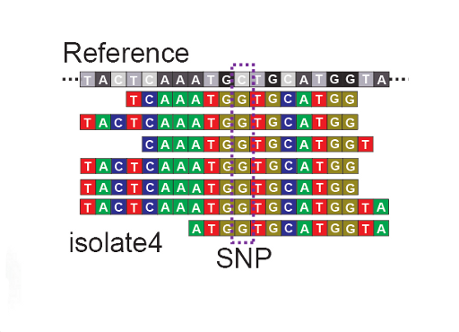 Determine the differences and similarities in the genetic code bewteen strains.
Determine the differences and similarities in the genetic code bewteen strains.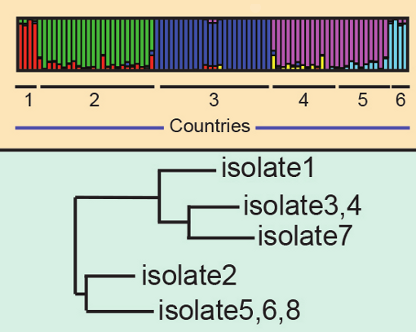 Use these similarities and differences to determine how related different strains of the pathogen are
Use these similarities and differences to determine how related different strains of the pathogen are
